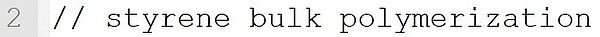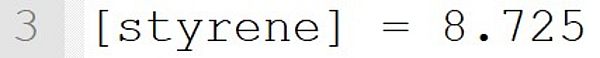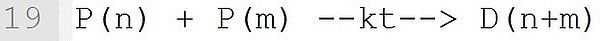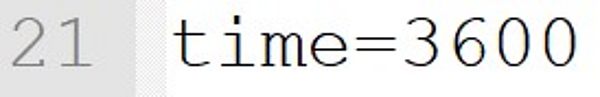Tutorial: model file Styrene.mcPolymer
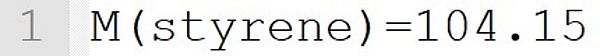
The molecular weight 104.15 g·mol-1 is assigned to the monomer styrene. This statement implicitly defines the species styrene as a monomer.
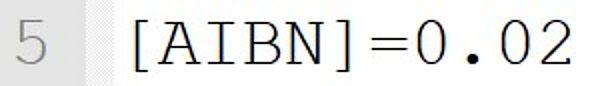
It implicitly defines a species called "AIBN". The concentration is set to 0.02 mol·L-1 for the beginning of the reaction.
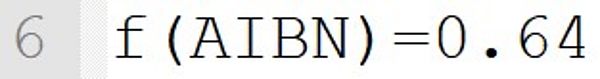
This statement implicitly defines the low molecular weight species AIBN as initiator. The initiator efficiency is 0.64 and can be chosen in the range of 0 to 1.
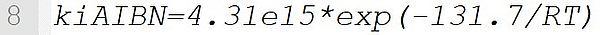
It defines a reaction rate coefficient with the pre-exponential factor 4.31e15 s-1 and the activation energy 131.7 kJ·mol-1.

It defines a reaction rate coefficient with the pre-exponential factor 1.341e10 L·mol-1·s-1 and the activation energy 14.34 kJ·mol-1. The spaces (e.g., before and after "*") generally do not affect the interpretation of the instruction.
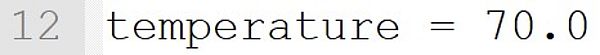
The temperature is set to 70 °C. If such an instruction is included in the model file, the temperature at the start of the program need not be entered on the command line. With an optional input on the command line (e.g., --Temperature = 60.0), this value is overruled and the simulation runs at 60 °C.
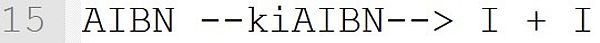
The initiator decomposition of AIBN becomes part of the kinetic model. The reaction is derived from the reaction template "initiator --k --> I + I" (see Table Reaction Templates). AIBN stands for the initiator, I for the primary radicals and kiAIBN for the reaction rate coefficient. The reaction template "initiator --k --> I + I" is only recognized correctly if the initiator has been assigned an initiator efficiency (see line 6). The primary radicals I are implicitly defined as low molecular weight species when used in a reaction. A separate definition (e.g., [I] = 0.0) is not necessary.
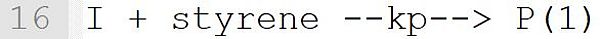
This reaction describes the initiation of a macro radical from the primary radical I with the monomer styrene. The concrete reaction was derived from Template "I + M --k--> P(1)" (see Table Reaction Templates). This template expects a monomer at position "M". Therefore, it is essential that the species styrene is assigned a molecular weight (see line 1). Only with this assignment the species styrene does get the property "monomer" and can be used in this reaction. The result is a macro radical P(n) with the current chain length n = 1. In formulating the reaction, the macroradical must be labeled as a macromolecule. In this initiation reaction, this is done by the name suffix (1).
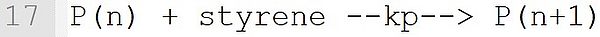
The propagation reaction formulated on this line is derived from Template P(n) + M --k--> P(n+1). Again, a monomer is expected at position "M". While the name of the macroradical is selectable by the user (e.g., P or MR), the extensions (n) in the educt and (n + 1) in the product should be used exactly as shown.
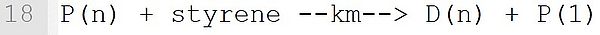
In this transfer reaction with the monomer styrene, a dead polymer D(n) and a new macroradical P with chain length 1 are formed. The associated reaction template is: P(n) + M --k--> D(n) + Q(1). It must be ensured that at the position "M" is a monomer and the macromolecular species with (n), (n) and (1) are marked exactly at the position shown.
![[Translate to English:] [Translate to English:]](/fileadmin/_processed_/7/3/csm_VRFB-2_7e0870b4fc.jpg)